Apostle 技术在许多国际领先的科研、临床实验室、公共卫生监测领域广泛应用。下面是一些在实时PCR(rtPCR)领域应用的案例。对于更多引用 Apostle 技术的文献、客户评价和反馈,请访问这里。
Safety and tolerability of AAV8 delivery of a broadly neutralizing antibody in adults living with HIV: a phase 1, dose-escalation trial.
Casazza et al. Nature Medicine. April 11, 2022
Adeno-associated viral vector-mediated transfer of DNA coding for broadly neutralizing anti-HIV antibodies (bnAbs) offers an alternative to attempting to induce protection by vaccination or by repeated infusions of bnAbs. In this study, we administered a recombinant bicistronic adeno-associated virus (AAV8) vector coding for both the light and heavy chains of the potent broadly neutralizing HIV-1 antibody VRC07 (AAV8-VRC07) to eight adults living with HIV. All participants remained on effective anti-retroviral therapy (viral load (VL) <50 copies per milliliter) throughout this phase 1, dose-escalation clinical trial (NCT03374202). AAV8-VRC07 was given at doses of 5 × 1010, 5 × 1011 and 2.5 × 1012 vector genomes per kilogram by intramuscular (IM) injection. Primary endpoints of this study were to assess the safety and tolerability of AAV8-VRC07; to determine the pharmacokinetics and immunogenicity of in vivo VRC07 production; and to describe the immune response directed against AAV8-VRC07 vector and its products. Secondary endpoints were to assess the clinical effects of AAV8-VRC07 on CD4 T cell count and VL and to assess the persistence of VRC07 produced in participants. In this cohort, IM injection of AAV8-VRC07 was safe and well tolerated. No clinically significant change in CD4 T cell count or VL occurred during the 1–3 years of follow-up reported here. In participants who received AAV8-VRC07, concentrations of VRC07 were increased 6 weeks (P = 0.008) and 52 weeks (P = 0.016) after IM injection of the product. All eight individuals produced measurable amounts of serum VRC07, with maximal VRC07 concentrations >1 µg ml−1 in three individuals. In four individuals, VRC07 serum concentrations remained stable near maximal concentration for up to 3 years of follow-up. In exploratory analyses, neutralizing activity of in vivo produced VRC07 was similar to that of in vitro produced VRC07. Three of eight participants showed a non-idiotypic anti-drug antibody (ADA) response directed against the Fab portion of VRC07. This ADA response appeared to decrease the production of serum VRC07 in two of these three participants. These data represent a proof of concept that adeno-associated viral vectors can durably produce biologically active, difficult-to-induce bnAbs in vivo, which could add valuable new tools to the fight against infectious diseases.
...
AAV8-VRC07 vector DNA quantitation. Plasma AAV8-VRC07 plasmid DNA was measured by extracting DNA from plasma, concentrating and then using a real-time PCR assay to measure a 103 base sequence spanning the junction of the IgG heavy chain sequence and F2A insert. DNA was extracted from serum using an Apostle MiniMax High Efficiency cfDNA Isolation Kit, following the manufacturer’s protocol with slight modification.
Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy.
Jin et al. PNAS February 2, 2021 118 (5) e2017421118; https://doi.org/10.1073/pnas.2017421118
Multiplex assays, involving the simultaneous use of multiple circulating tumor DNA (ctDNA) markers, can improve the performance of liquid biopsies so that they are highly predictive of cancer recurrence. We have developed a single-tube methylation-specific quantitative PCR assay (mqMSP) that uses 10 different methylation markers and is capable of quantitative analysis of plasma samples with as little as 0.05% tumor DNA. In a cohort of 179 plasma samples from colorectal cancer (CRC) patients, adenoma patients, and healthy controls, the sensitivity and specificity of the mqMSP assay were 84.9% and 83.3%, respectively. In a head-to-head comparative study, the mqMSP assay also performed better for detecting early-stage (stage I and II) and premalignant polyps than a published SEPT9 assay. In an independent longitudinal cohort of 182 plasma samples (preoperative, postoperative, and follow-up) from 82 CRC patients, the mqMSP assay detected ctDNA in 73 (89.0%) of the preoperative plasma samples. Postoperative detection of ctDNA (within 2 wk of surgery) identified 11 of the 20 recurrence patients and was associated with poorer recurrence-free survival (hazard ratio, 4.20; P = 0.0005). With subsequent longitudinal monitoring, 14 patients (70%) had detectable ctDNA before recurrence, with a median lead time of 8.0 mo earlier than seen with radiologic imaging. The mqMSP assay is cost-effective and easily implementable for routine clinical monitoring of CRC recurrence, which can lead to better patient management after surgery.
Plasma DNA extraction was performed using 2 to 5 mL of plasma with the Apostle MiniMax High-Efficiency cfDNA Isolation Kit, according to the product manual.
Jin et al. Clinical Chemistry and Laboratory Medicine (CCLM). 2021; 59(1): 91–99
Objectives - Colorectal cancer (CRC) screening using stool samples is now in routine use where tumor DNA methylation analysis for leading markers such as NDRG4 and SDC2 is an integral part of the test. However, processing stool samples for reproducible and efficient extraction of human genomic DNA remains a bottleneck for further research into better biomarkers and assays.
Methods - We systematically evaluated several factors involved in the processing of stool samples and extraction of DNA. These factors include: stool processing (solid and homogenized samples), preparation of DNA from supernatant and pellets, and DNA extraction with column and magnetic beads-based methods. Furthermore, SDC2 and NDRG4 methylation levels were used to evaluate the clinical performance of the optimal protocol.
Results - The yield of total and human genomic DNA (hgDNA) was not reproducible when solid stool scraping is used, possibly due to sampling variations. More reproducible results were obtained from homogenized stool samples. Magnetic beads-based DNA extraction using the supernatant from the homogenized stool was chosen for further analysis due to better reproducibility, higher hgDNA yield, lower non-hgDNA background, and the potential for automation. With this protocol, a combination of SDC2 and NDRG4 methylation signals with a linear regression model achieved a sensitivity and specificity of 81.82 and 93.75%, respectively.
Conclusions - Through the systematic evaluation of different stool processing and DNA extraction methods, we established a reproducible protocol for analyzing tumor DNA methylation markers in stool samples for colorectal cancer screening.
(Methods section) For magnetic beads-based method, Apostle Stool gDNA Isolation Kit (APOSTLE) was used according to the manufacturer’s instructions. Either 0.2 g pellets or 0.2 mL supernatant from homogenized stool was mixed with 1 mL lysis buffer (APOSTLE) for DNA extractions.
BGI’s Three Complementary Kits of Colorectal Cancer Testing Have Been CE Marked.
BGI. July 13, 2021
BGI Genomics announces that its Stool Sample Collection Kit, DNA Isolation Kit together with Sample Pretreatment Kit for Methylation Detection have been CE marked.
All three kits are used in conjunction with the previously CE marked Colorectal Cancer Testing Product which can detect the methylation of SDC2, ADHFE1 and PPP2R5C genes in human fecal samples.
According to the Global Cancer 2020 (GLOBOCAN) statistics, there are about 19.3 million new cases of colorectal cancer each year, accounting for 10 percent of all new cancer cases. About 935,000 colorectal cancer deaths occur each year, accounting for 9.4 percent of all cancer deaths.
So far, BGI has obtained CE mark for all four products used in the colorectal cancer detection workflow, from sample collection, DNA extraction, DNA pre-treatment to methylation detection, providing customers with reliable and standardized reagents and services.
(Note: Apostle is the Original Equipment Manufacturer or OEM for the Stool DNA Isolation Kit mentioned in this news. Apostle 's branded product is called Apostle MiniGenomics Stool Fast Kit.)
Gang Peng,* Yibo Xi,* Chiara Bellini, Kien Pham, Zhen W Zhuang, Qin Yan, Man Jia, Guilin Wang, Lingeng Lu, Moon-Shong Tang, Hongyu Zhao, and He Wang American Journal of Cancer Research. Volume 12, Issue 8, August 2022, Pages 3679–3692.
Abstract
Epigenomic-wide DNA methylation profiling holds the potential to reflect both electronic cigarette exposure-associated risks and individual poor health outcomes. However, a systemic study in animals or humans is still lacking. Using the Infinium Mouse Methylation BeadChip, we examined the DNA methylation status of white blood cells in male ApoE-/- mice after 14 weeks of electronic cigarette exposure with the InExpose system (2 hr/day, 5 days/week, 50% PG and 50% VG) with low (6 mg/ml) and high (36 mg/ml) nicotine concentrations. Our results indicate that electronic cigarette aerosol inhalation induces significant alteration of 8,985 CpGs in a dose-dependent manner (FDR<0.05); 7,389 (82.2%) of the CpG sites are annotated with known genes. Among the top 6 significant CpG sites (P-value<1e-8), 4 CpG sites are located in the known genes, and most (3/5) of these genes have been related to cigarette smoking. The other two CpGs are close to/associated with the Phc2 gene that was recently linked to smoking in a transcriptome-wide associations study. Furthermore, the gene set enrichment analysis highlights the activation of MAPK and 4 cardiomyocyte/cardiomyopathy-related signaling pathways (including adrenergic signaling in cardiomyocytes and arrhythmogenic right ventricular cardiomyopathy) following repeated electronic cigarette use. The MAPK pathway activation correlates well with our finding of increased cytokine mRNA expression after electronic cigarette exposure in the same batch of mice. Interestingly, two pathways related to mitochondrial activities, namely mitochondrial gene expression and mitochondrial translation, are also activated after electronic cigarette exposure. Elucidating the relationship between these pathways and the increased circulating mitochondrial DNA observed here will provide further insight into the cell-damaging effects of prolonged inhalation of e-cigarette aerosols.
(Methods section) Plasma cfDNA and mtDNA/nDNA ratio and oxidization of cell-free DNA
For measurement of in vivo plasma mtDNA/nDNA ratio, mice plasma was first subjected to cell-free DNA (cfDNA) extraction using Apostle minimax cfDNA extraction kit (ApostleBio, CA, US).
Interview with Lauryn Massic, Association of Public Health Laboratories infectious disease fellow at the Nevada State Public Health Laboratory
Lauryn Massic, Association of Public Health Laboratories infectious diseases fellow, shares how breakthrough technologies from Ceres Nanosciences and Apostle enables SARS-CoV-2 wastewater surveillance results that are efficient and accurate.
Please share with us more about your workflow and your metrics for success.
LM: Our goal in setting up our workflow was to have a turnaround time of less than one day for detection and quantification of the virus in wastewater samples. We wanted to be able to test wastewater from each community facility at least three times per week and to test the campus dorms every day of the week.
The workflow we follow in the lab starts with the use of Ceres Nanosciences’ Nanotrap Magnetic Virus Particles on the Apostle MagTouch 2000 for virus concentration from the wastewater. This is followed by RNA extraction using ThermoFisher and Apostle Bio reagents on the MagTouch 1000. We are analyzing the RNA using the Promega SARS-CoV -2 Wastewater RT-qPCR kit and are sequencing extracted RNA using Illumina short read sequencing on a MiniSeq.
How has the Apostle automation accelerated your testing capabilities?
LM: With Apostle automation, my team and I have been able to develop a method to detect and quantify SARS-CoV-2 in wastewater samples, while also having the ability to sequence viral RNA from the wastewater samples. We can accomplish the detection and quantification portion all in the span of a day, and the hands- free time of the automated process gives us the ability to complete other wastewater-related tasks.
Rapid repeat infection of SARS-CoV-2 by two highly distinct delta-lineage viruses.
Andrew J. Gorzalski , Christina Boyles, Victoria Sepcic, Subhash Verma, Joel Sevinsky, Kevin Libuit, Stephanie Van Hooser, Mark W. Pandori. Diagnostic Microbiology and Infectious Disease. Volume 104, Issue 1, September 2022, 115747; https://doi.org/10.1016/j.diagmicrobio.2022.115747
An instance of sequential infection of an individual with, firstly, the Delta variant and secondly a Delta-sub-lineage has been identified. The individual was found positive for the AY.26 lineage 22 days after being found positive for the Delta [B.1.617.2] variant. The viruses associated with the cases showed dramatic genomic difference, including 31 changes that resulted in deletions or amino acid substitutions. Seven of these differences were observed in the Spike protein. The patient in question was between 30 and 35 years old and had no underlying health conditions. Though singular, this case illustrates the possibility that infection with the Delta variant may not itself be fully protective against a population of SARS-CoV-2 variants that are becoming increasingly diverse.
Nucleic acid extractions were performed by Apostle MagTouch Nucleic Acid Extraction Automation Systems [Apostle Inc, San Jose, CA].
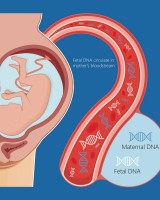
The SneakPeek Early Gender DNA Test
SneakPeek by Gateway Genomics was founded with the goal to make DNA-based prenatal and pediatric information accessible and affordable for parents everywhere.
The SneakPeek Early Gender DNA Test is designed to specifically focus on fetal sex and return just one answer – male or female. This enables the test to be run on magnitudes smaller volumes of blood than the typical non-invasive prenatal test. When maternal blood samples arrive at SneakPeek Labs, extracted cell-free fetal DNA is run through real-time quantitative PCR to detect with a sensitivity down to a single Y chromosome. If Y chromosomes are found, the result is a boy. If they are absent, the baby is a girl. The test is run in 3 hours with 99.9% accuracy¹.
The SneakPeek Early Traits DNA Test lets parents find out what their infant or child will look like as an adult, predicted nutrition levels and sleep behavior. The DNA collection process is simple with a rub of the inner cheek using a swab, no blood samples are required for this test making it easy for parents. When the DNA samples arrive at SneakPeek Labs, genotyping method is used to analyze the differences in DNA and determine which traits an individual may have as a result.
(Note: Apostle is a technological product provider to SneakPeek by Gateway Genomics).
¹In a recent large-scale study, SneakPeek accurately determined fetal sex in 99.9% of 1,029 pregnant women between 7-37 weeks gestational age. In a separate clinical study run in 2021, SneakPeek accurately determined fetal sex in 75 out of 75 pregnant women at 7 weeks into pregnancy.
对于更多应用和引用 Apostle 技术的文献、客户评价和反馈,请访问这里。